DNAJupyter
Enable users to use JupyterLab smoothly in DNAnexus platform.
An important feature launched in Jan 2019 on DNAnexus Apollo platform .
Type
Web app features resulted from my internship at DNAnexus
Time
2018.7-2018.8
Teammates
Sang Soo Jun (PM) and more stakeholders
My Role
UX Designer
Skills
Interaction design / colaboration with a multi-functional team
Tools
Sketch / InVision
BACKGROUND & GOAL
DNAnexus users use JupyterLab frequently:
DNAnexus helps users store their tremendous-sized genomic data and run related apps to manipulate / analyze the data.
JupyterLab enable users develop apps to handel specific data on their own.
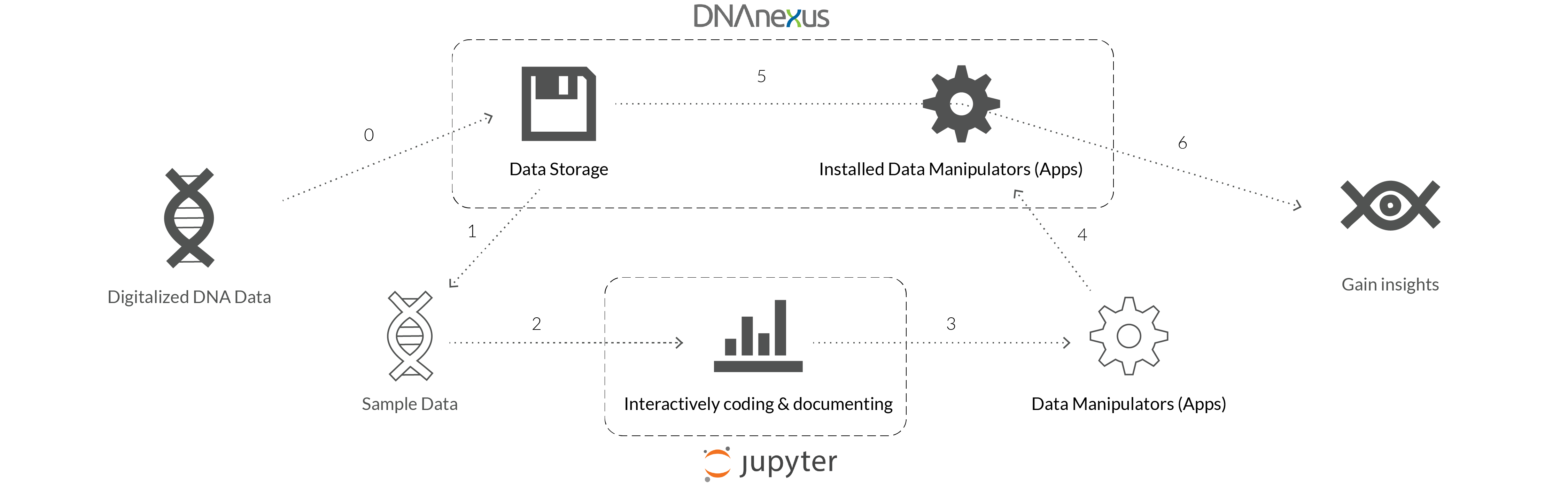
The pain point of the users:
"As a Bio-statistician, I use DNAnexus to store my data and the its apps to manipulate the data. But in order to actively interact with the data,
I have to switch to another webapp, JupyterLab..."
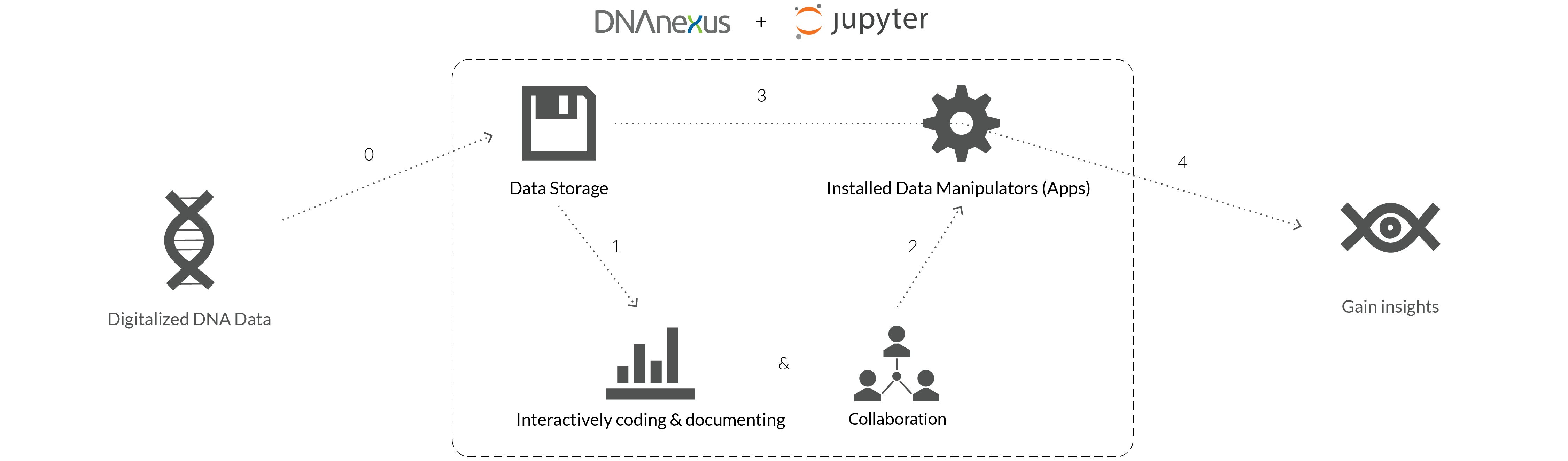
The Goal:
"As a Bio-statistician, I will use JupyterLab inside DNAnexus so that I won't have to switch back and force to work on my research data."
DESIGN PROCESS
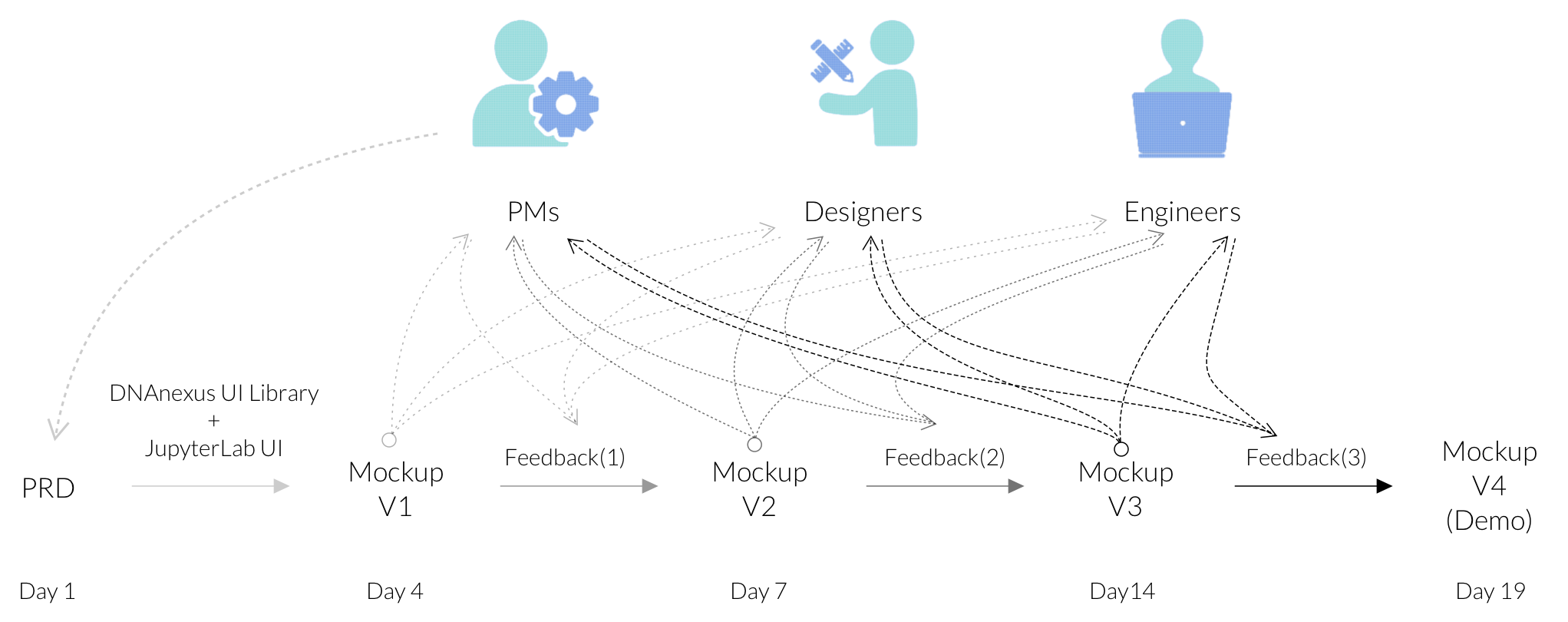
Gather feedback and Iterate:
For this project, I started with handing in PRD with wireframes done by PMs. Without practically easy access to the direct users, the collaborations with cross-functional teams and the feedback from stakeholders were foundamental to the iterations and process.
DESIGN DETAILS
The whole project involved 54 pages dealing with more than 12 design problems in the flow.
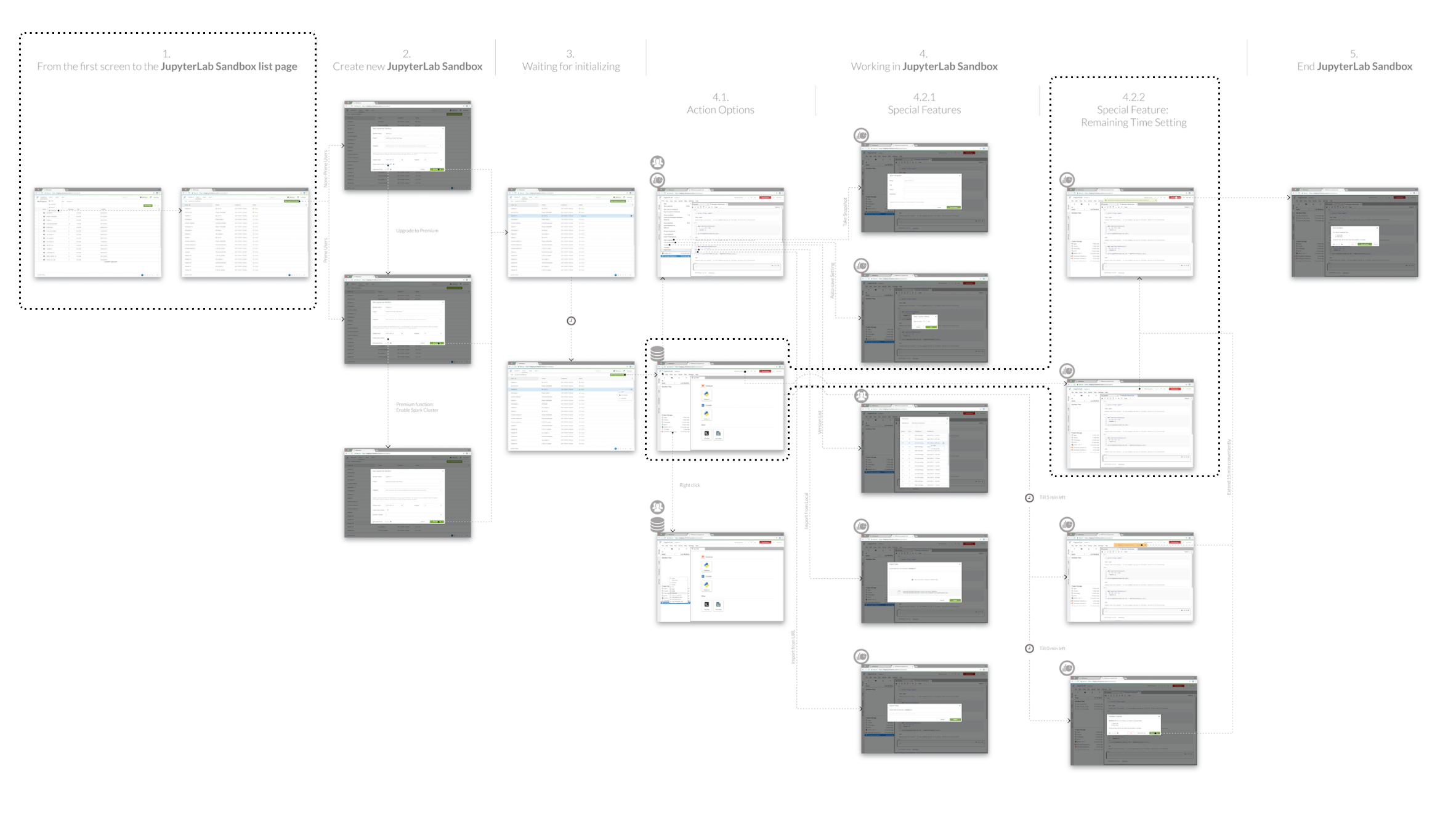
Here I would showcase 2 examples of design problems (result in the marked areas in the sitemap) on how I approach them.
Design problem example 1: Where to start?
step 0: Wireframe from PRD
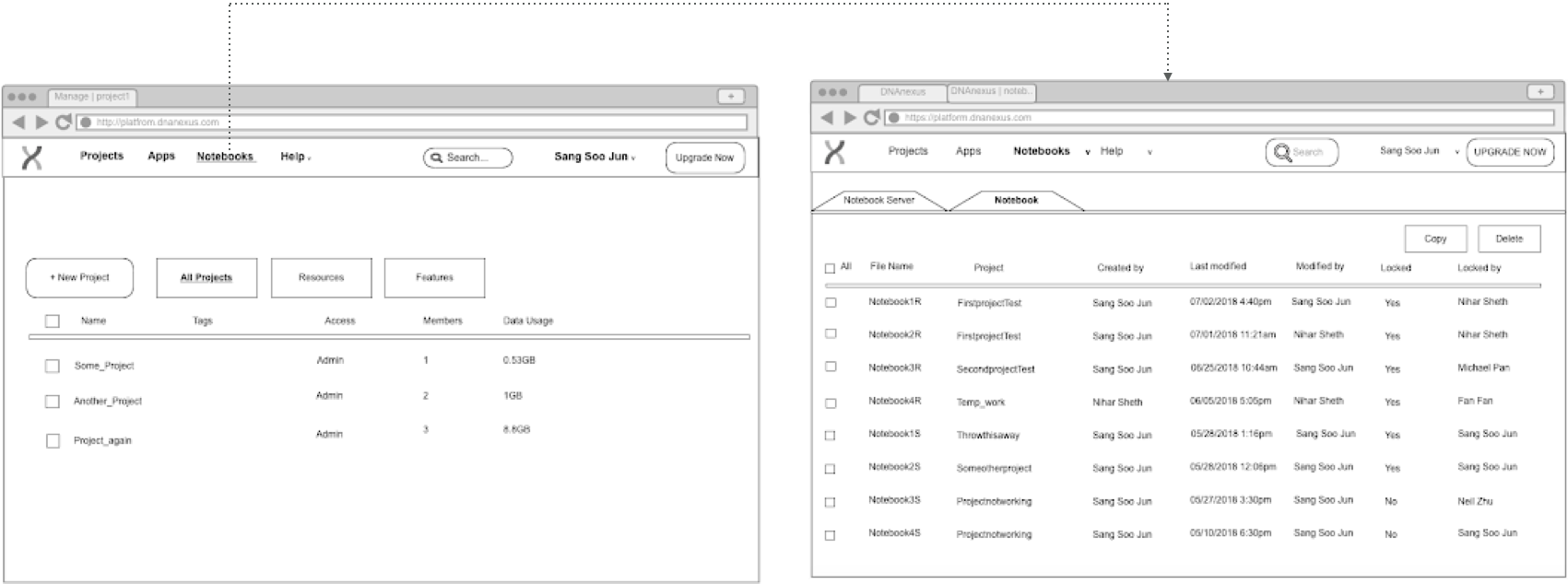
step 1: Ideation and comparison
Original Version (translated from PRD):

Proposed Version 1 (grouping tabs: "JupyterLab" and "Apps" into "Tools"):

Proposed Version 2 (combining segments: "Server" and "Jupyter Files"):
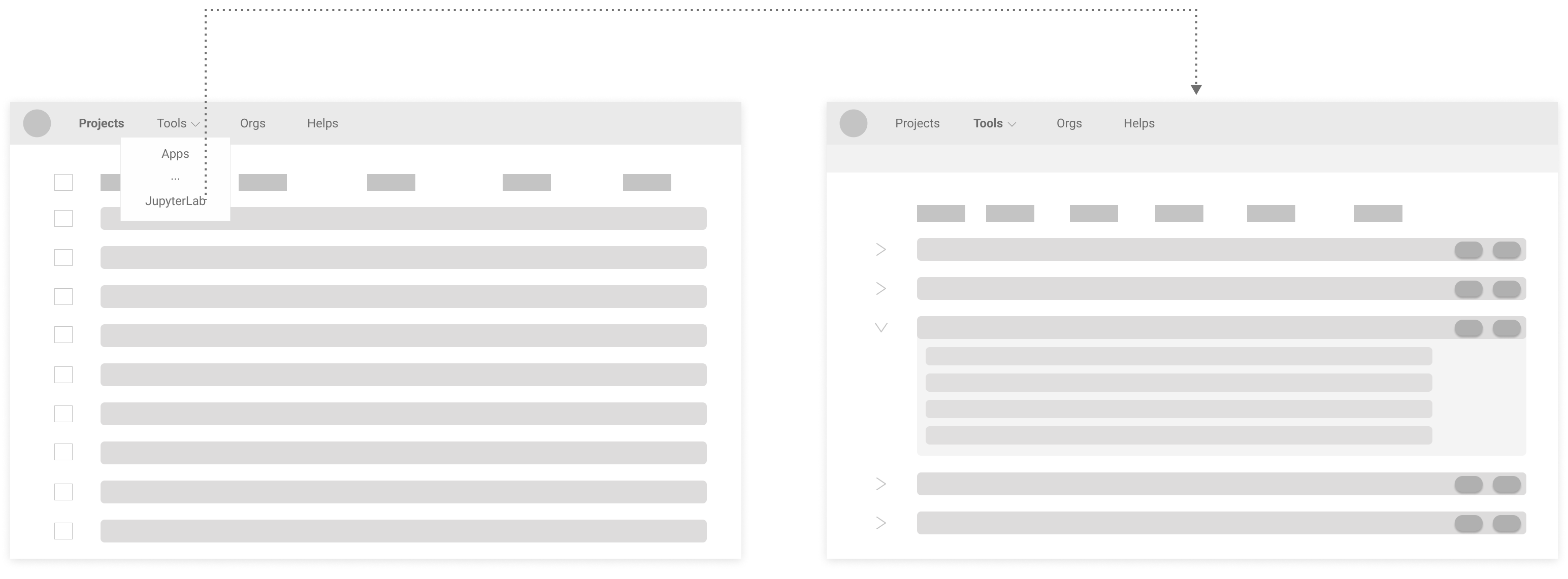
Based on discussions with stakeholders, I decided to give the Proposed Version 2 a try because of its conciseness and clarity.
step 2: Mockup prototype to gather feedback
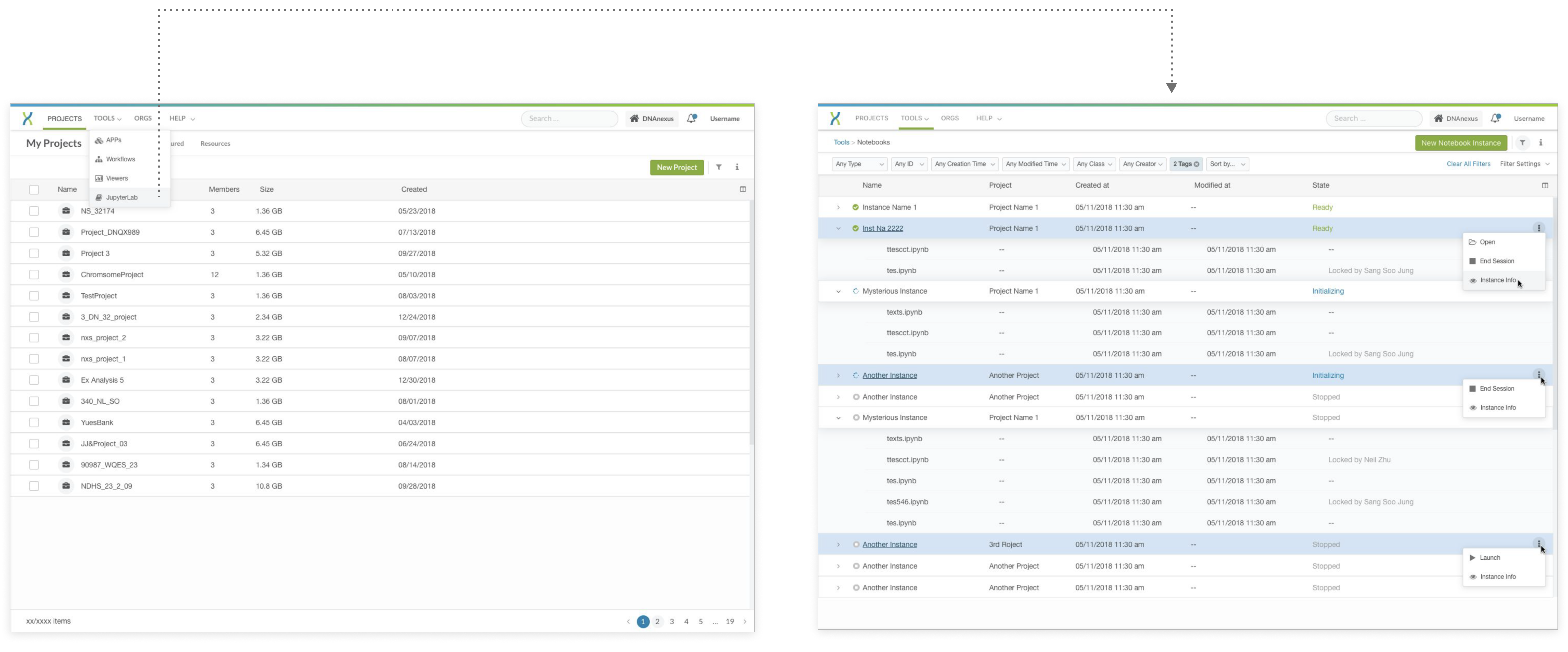
Feedback from users and stakeholders:
Users didn’t have the need to see what files are associated with what server, since they are multiple-to-multiple, this expansable list created more confusions to them.
Users didn’t need to see a standalone Jupyter files list, since the files can be listed in the storage like other files, which will be linked to the server under the same project.
Engineers pointed out that there were no server info available for the initiating and ended servers.
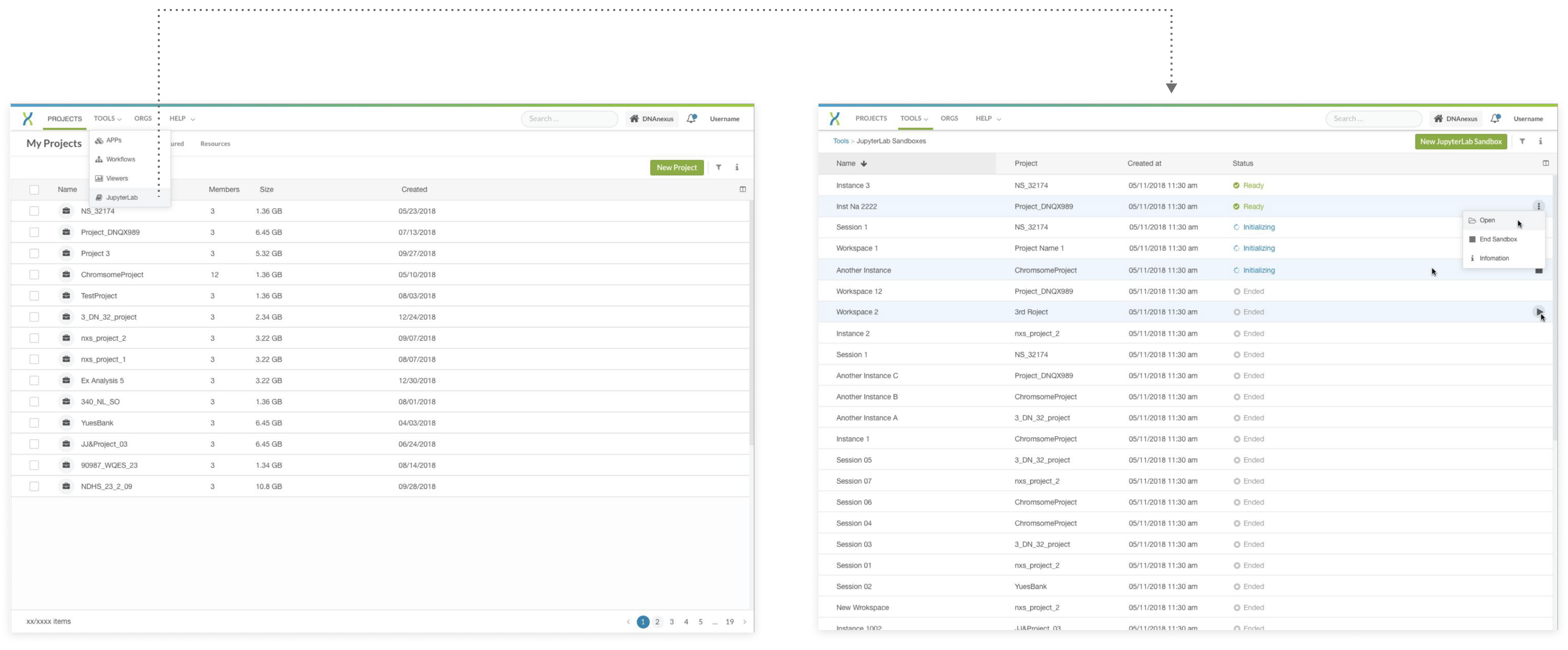
step 3: Iteration based on the feedback
Design problem example 2: Running time management?
Why does this matter:
Running server costs money.
Not every user has the habit to stop servers whenever tasks are done, which results in financial wastes.
step 0: Wireframe from PRD
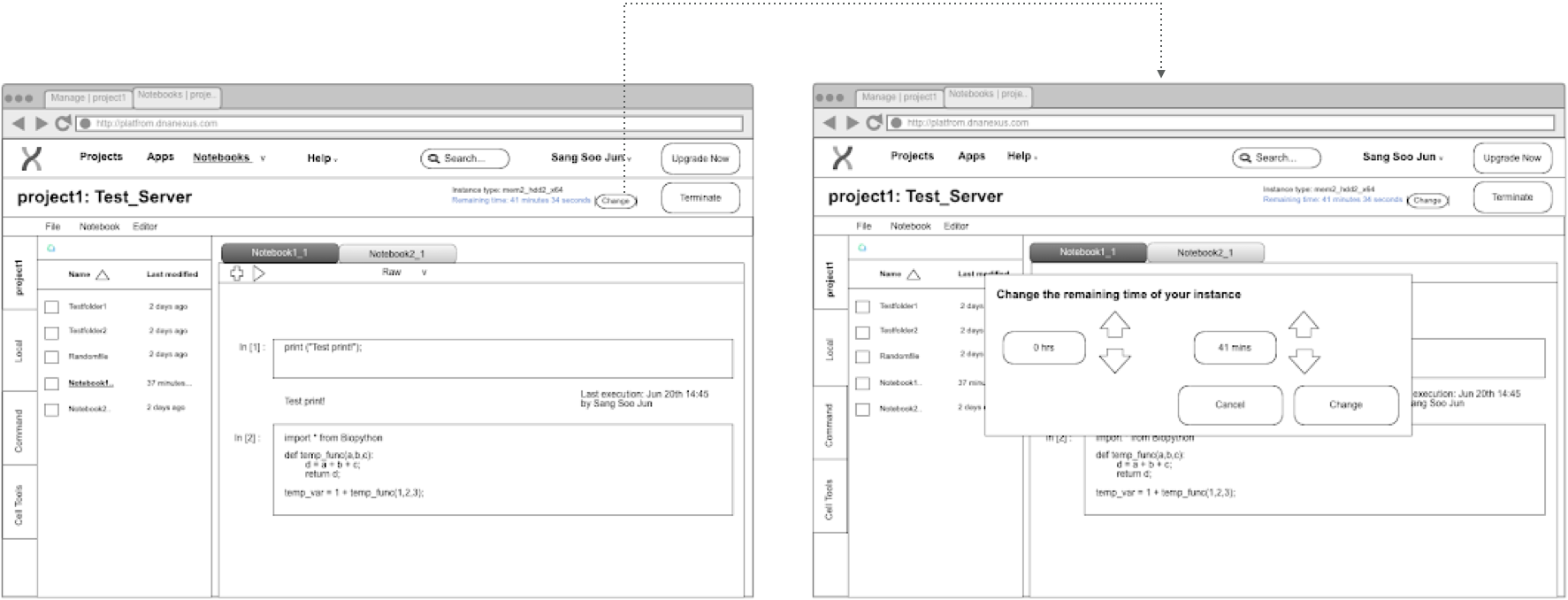
step 1: Ideation and comparison with usability test
Original Version (translated from PRD):
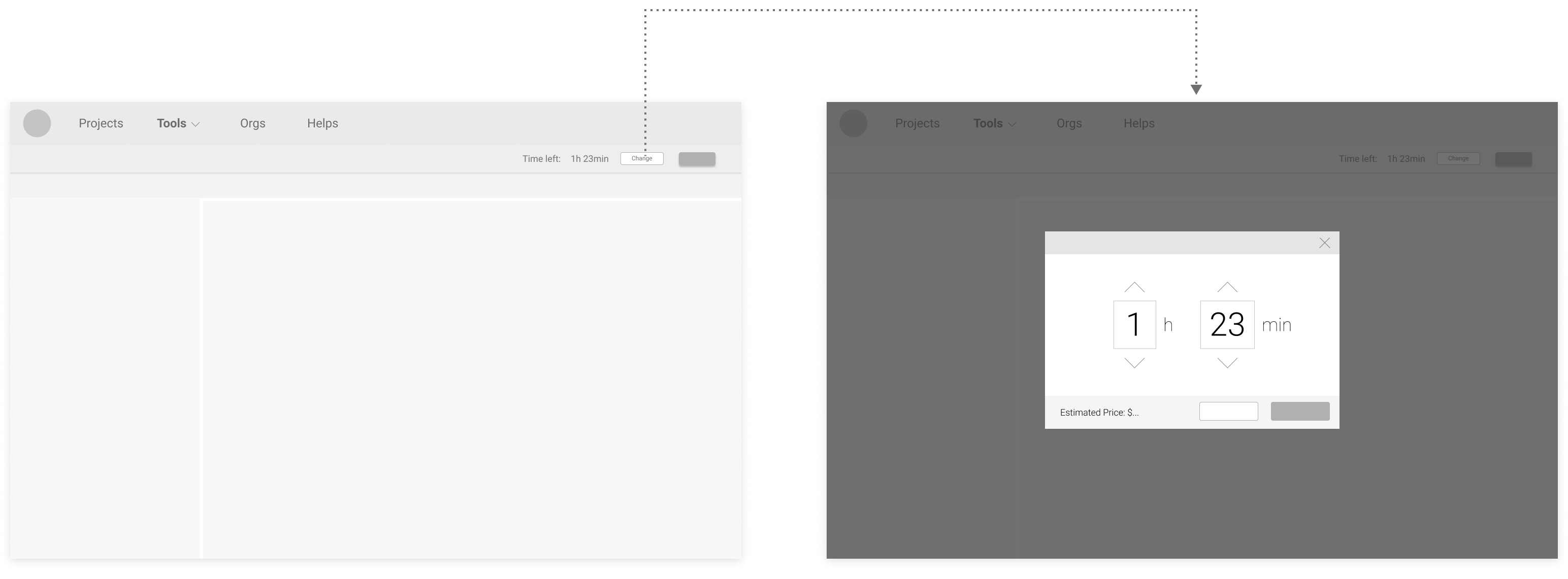
Proposed Version 1 (with preset options of time spans):
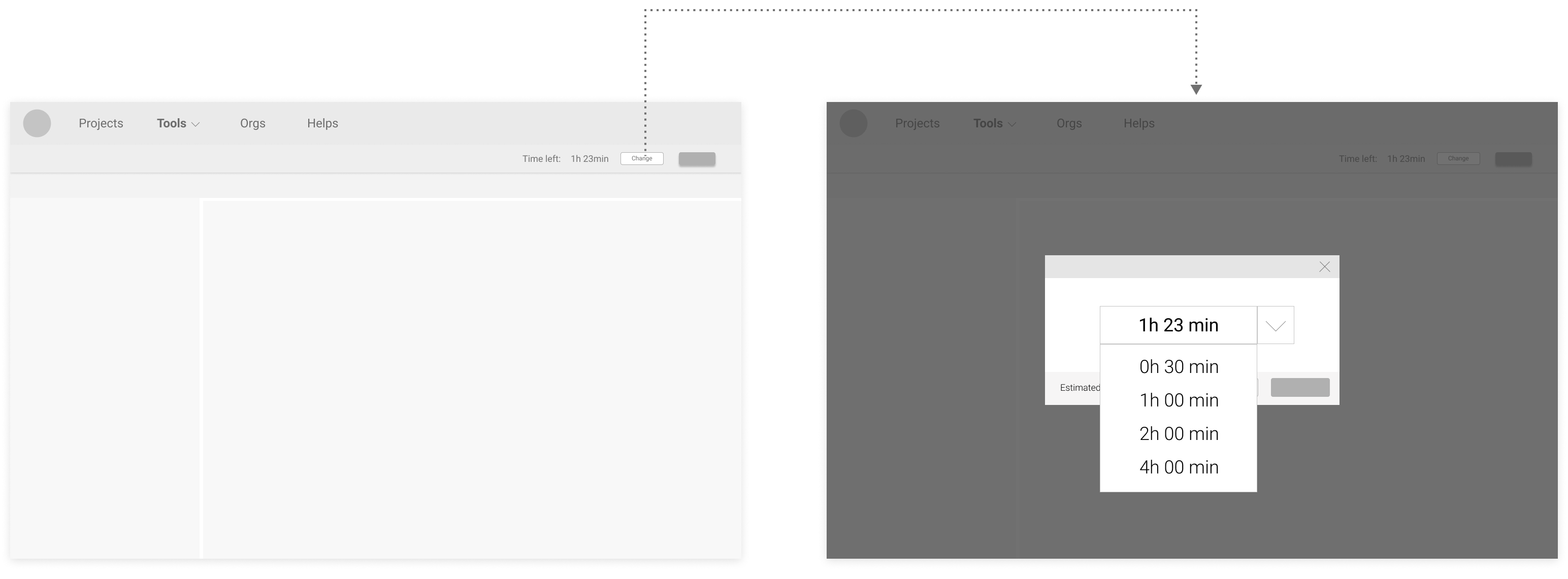
Proposed Version 2 (options of time spans at the typefield):
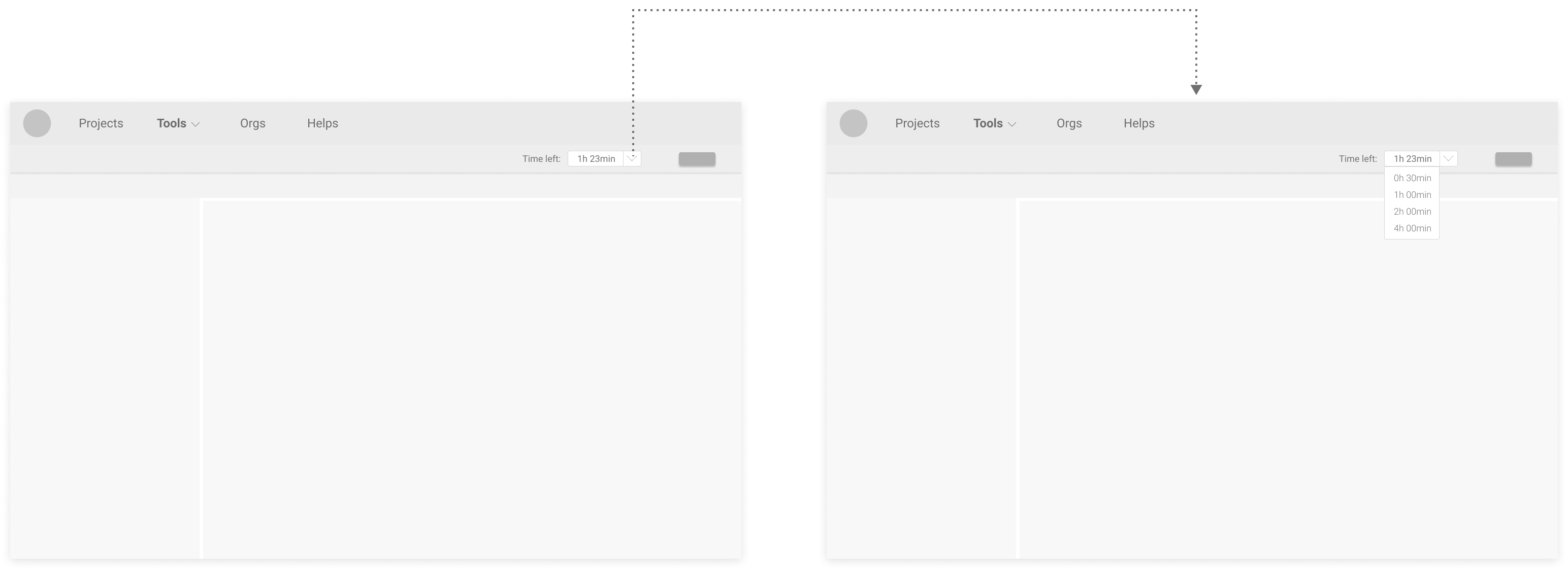
Proposed Version 3 (direct change at the typefield):
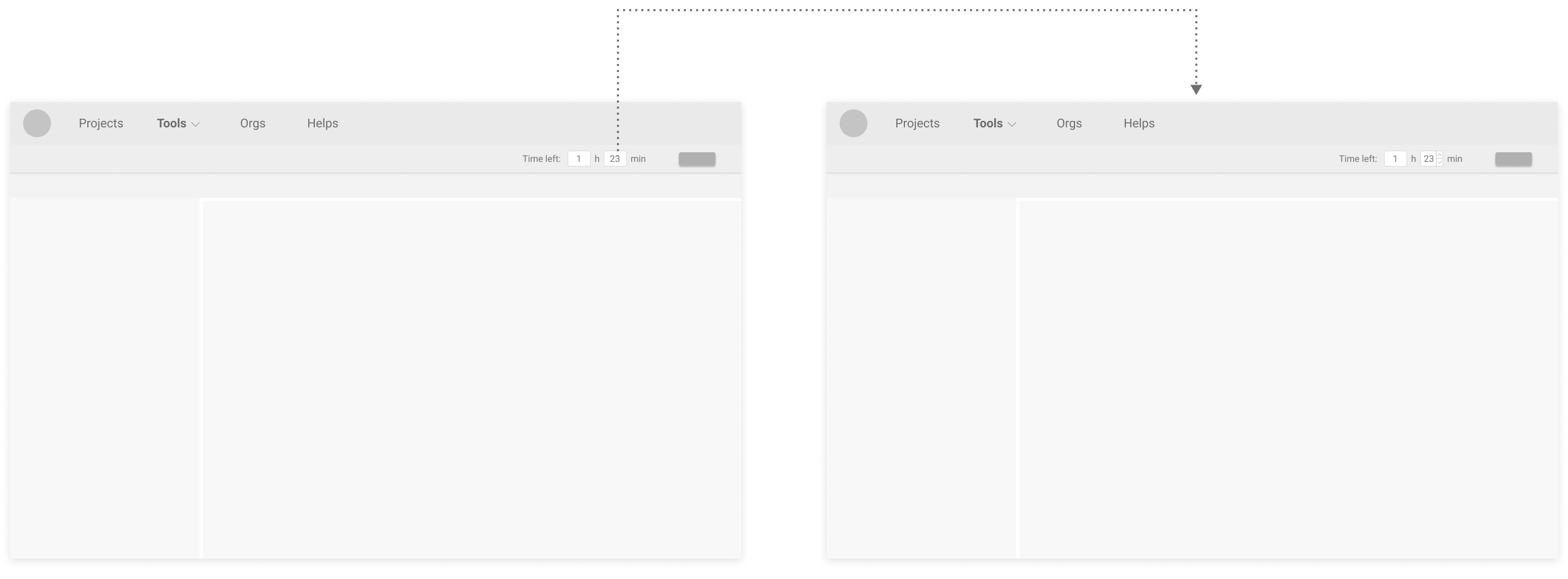
Based on feedback following a simple usability test:
Users preferred modifying time directly at the typefield, like in Proposed Version 2 & 3, because it feels more efficient.
Although many users liked having preset choices like in Proposed Version 1 & 2, no consensus on what options should be there was made.
Users also liked the flexibility in Proposed Version 3
Therefore, I decided to go develope the design on Proposed Version 3
step 2: Focus on the detailed interaction
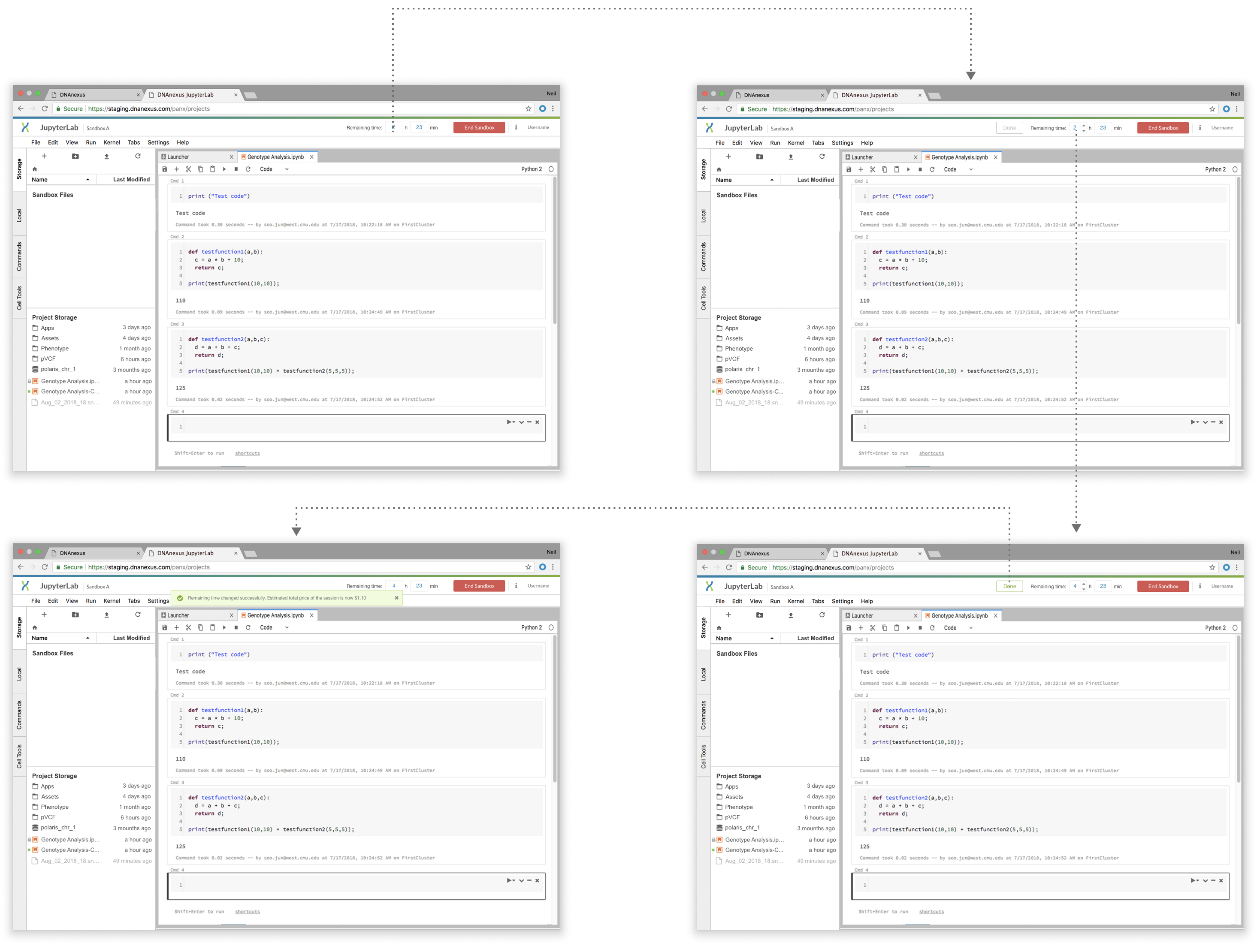
The usability test after this version revealed no usability issues, and engineers agreed on the doability of this flow.
SOLUTION DEMO
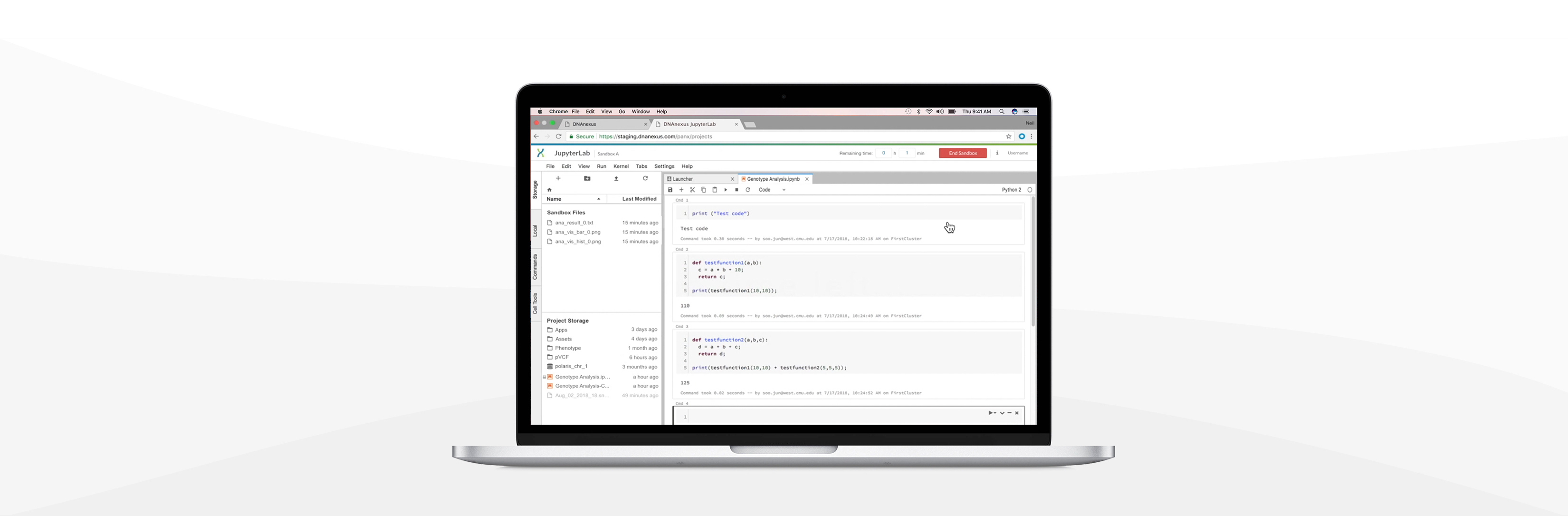
Example 1: Bob (a user) starts using Jupyterlab in DNAnexus.
Example 2: Bob modifies the running time at multiple moments.
REFELECTIONS
TAKEAWAY
-
PRD is a start point to work with PM on further user research and validation rather before diving into design details.
-
"Love the problem, not the solution" - do not stick to one solution, but ideate more alternatives for the problem and prioritize from there.
-
Do not underestimate seemingly small-scoped features, which can be valuable and more complicated than they seem.
OUTCOME
DNAnexus Apollo was launched in Jan 2019, and one of its 6 foundamental features is "Empower biologists to explore data", which basically talks about "DNAJupyter".
